Estimate manifold dimensionality with LID
Sunday December 27, 2020
Local Intrinsic Dimensionality (LID) is based on assuming locally uniform density and using the geometric idea that in \( D \) dimensions the volume of a ball with radius \( r \) goes as \( r^D \). With an outer radius \( r_{\text{bound}} \) from some point and a distance \( r \lt r_{\text{bound}} \) to a neighboring point, the maximum likelihood estimator of LID is \( 1 / \log{\left( r_{\text{bound}}/r \right)} \), and multiple estimates are averaged with the harmonic mean.
For example, Swiss roll data curves through three dimensions but only on a two-dimensional manifold. LID for the three-dimensional Swiss roll data is about two, which is informative.
- Calculating LID with Python
- Mathematical derivation of LID
- Caveats
- LID in the literature
- Topological extensions
Calculating LID with Python
The maximum likelihood estimator of Local Intrinsic Dimensionality is \( 1 / \log{\left( r_{\text{bound}}/r \right)} \).
import math
def lid(radius, radius_bound):
return 1 / math.log(radius_bound / radius)The radii will be distances measured from data using Euclidean distance.
def euclidean(xs, ys): # Euclidean distance
return math.sqrt(sum((x - y)**2 for x, y in zip(xs, ys)))To average multiple estimates of dimensionality, use the harmonic mean.
def harmonic_mean(numbers):
return len(numbers) / sum(1 / number for number in numbers)Filter out zeros (such as the distance from a point to itself).
def positive(numbers):
return [number for number in numbers if number > 0]For a point in some data, find the distances to its \( k+1 \) nearest neighbors. The greatest of these is \( r_{\text{bound}} \), and the estimates based on the \( k \) nearest neighbors are averaged.
def neighbors_lid(origin, points, k=1, distance=euclidean):
radii = sorted(positive([distance(origin, point) for point in points]))
return harmonic_mean([lid(radius, radii[k]) for radius in radii[:k]])In a cloud of points
A uniformly random cloud of points in \( D \) dimensions has dimensionality equal to \( D \).
import random
def cloud(dimensions, count=100, bounds=(-1, 1)):
return [[random.uniform(*bounds) for _ in range(dimensions)]
for _ in range(count)]LID can be calculated from anywhere in the cloud, such as the origin, as in this seven-dimensional example.
neighbors_lid([0]*7, cloud(7))
## 6.851551579378687These estimates are very noisy, depending on the particular random points in the cloud; it's very likely that a given run won't be so close to seven. Repeating the experiment many times is more reassuring.
harmonic_mean([neighbors_lid([0]*7, cloud(7)) for _ in range(1000)])
## 7.006859728132216For a Swiss roll dataset
To estimate a dataset's dimensionality, average LID from each point in the dataset.
def data_lid(points, k=1, distance=euclidean):
return harmonic_mean([neighbors_lid(point, points, k=k, distance=distance)
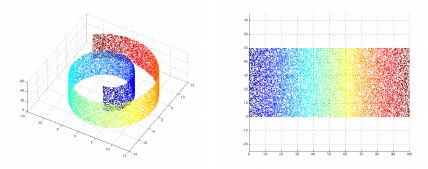
for point in points])Swiss roll data, first introduced in the Isomap paper, is a two-dimensional manifold embedded in three dimensions. It's not terribly hard to make from scratch, but the scikit-learn implementation is convenient.
import sklearn.datasets
roll, _ = sklearn.datasets.make_swiss_roll()
data_lid(roll)
## 2.0706005383894444The code here is also available in a notebook with slightly more technical commentary.
Mathematical derivation of LID
Local Intrinsic Dimensionality arises from considering radius from a given point to another point nearer than \( r_{\text{bound}} \) as a random variable \( R \) which takes a specific value \( r \).
Cumulative Distribution Function (CDF) of radius
With uniform density, the probability of encountering a point is proportional to volume, and the volume of a radius \( r \) ball in \( D \) dimensions is proportional to \( r^D \), giving the CDF.
\[ P(R<r) = \frac{r^D}{r_{\text{bound}}^D} = \left(\frac{r}{r_{\text{bound}}}\right)^D \]
Probability Density Function (PDF) of radius
The PDF is the derivative of the CDF with respect to \( r \).
\[ P(R=r) = D \left( \frac{r}{r_{\text{bound}}} \right)^{D-1} \]
Expected Value of radius
Integrating the PDF times \( r \) from 0 to \( r_{\text{bound}} \) gives the expected value of \( R \), which is \( \frac{D}{D+1} r_{\text{bound}} \). This isn't terribly important to the derivation here, but it shows that as dimensionality increases, points are likely to be closer and closer to the outer radius. This is an aspect of the curse of dimensionality and the phenomenon that LID is based on.
Maximum Likelihood Estimate of dimension
For a given measured \( r \) within \( r_{\text{bound}} \), some dimensionality \( D \) maximizes the likelihood of that observation. The probability density function is a likelihood, and the log of that function has the same maximum where both derivatives are zero. Setting the derivative with respect to \( D \) of the natural log likelihood equal to zero gives a simple solution.
\[ \log{\left( \frac{r}{r_{\text{bound}}} \right)} + \frac{1}{D} = 0 \]
\[ D = \frac{1}{\log{ \left( \frac{r_{\text{bound}}}{r} \right)}} \]
This choice of natural log is not arbitrary, because a derivative is taken; using a different log base would introduce an additional constant factor.
Harmonic mean for dimensionality
With two observed radii \( r_1 \) and \( r_2 \), the joint probability is two (PDF) likelihoods multiplied together, and the log likelihood sums. Maximizing gives the harmonic mean of two individual estimates.
\[ \log{\left( \frac{r_1}{r_{\text{bound}}} \right)} + \frac{1}{D} + \log{\left( \frac{r_2}{r_{\text{bound}}} \right)} + \frac{1}{D} = 0 \]
\[ D = \frac{2}{\log{\left( \frac{r_{\text{bound}}}{r_1} \right)} + \log{\left( \frac{r_{\text{bound}}}{r_2} \right)}} \]
The same thing happens for any number of measurements; the harmonic mean is the appropriate way to average estimates of dimensionality.
The harmonic mean is typically appropriate for averaging rates, so its appearance here suggests thinking of dimension as a rate. Perhaps dimension is the rate at which "space" becomes available, or entropy becomes possible, per distance traveled.
Caveats
It's very common for distance to not really make sense in a dataset. Scaling of variables can be very important, for example (not to mention categorical variables).
Even with a meaningful distance, in high dimensions, "nearest neighbors" may not be meaningful.
Euclidean distance is assumed here, but other distances may also be worth considering. There is also an angle-based approach to estimating LID.
LID in the literature
A common reference is Levina and Bickel, "Maximum Likelihood Estimation of Intrinsic Dimension." Their derivation is slightly different, but the result is the same: their Equation 8 is the MLE above in different notation.
Levina and Bickel suggest an adjustment to the harmonic mean when \( k \rightarrow \infty \). In practice it may be better to keep \( k \) small in the interest of maintaining locality and not adjust.
Another form of the MLE sometimes appears (e.g. 1, 2) but is not recommended.
\[ \widehat{LID}(x) = -\left( \frac{1}{k} \sum_{i=1}^k{ \log{ \frac{r_i(x)}{r_k(x)} } } \right)^{-1} \]
In this form, \( r_i(x) \) is the distance to the \( i\text{-th} \) nearest neighbor of the point \( x \), and \( r_k(x) \) is the maximum of these distances. Here the \( k \) neighbors include the farthest one, defining \( r_k(x) \), which is elsewhere called \( r_{\text{bound}} \). The effect of this inclusion is to add a \( \log(1) = 0 \) term to the sum (which would not be meaningful as an individual estimate) or equivalently to average \( k-1 \) estimates as if there were \( k \). With two nearest neighbors, this will incorrectly double the estimate, and it will always inflate it to some degree.
In addition to the arithmetic mean, sometimes a regression approach is used to combine estimates of dimensionality. Facco et al. apply such an approach, noting and dropping large outliers in order to get more robust estimates. The harmonic mean (itself a kind of robust regression) is more appropriate.
Topological extensions
Topological approaches use nearest neighbors to turn point-like data into graph-like data (okay, simplicial complexes). UMAP is a popular modern technique based on this idea.
In Isomap, a graph enforces geodesic distances, staying on the manifold via neighbors connected in the graph. For estimating dimensionality, a graph lets you avoid polluting the set of nearest neighbors with points from actually distant parts of the manifold (imagine jumping between sheets in Swiss roll data, for example). Some work on dimensionality (e.g. A, B) involves this approach.
Stephen Wolfram has some topological ideas about physics, including a very literal interpretation of dimensionality being the rate at which traveling a distance gives you access to more space.